The FlowCam Student Equipment Grant has helped many graduate students complete their research. Beyond being awarded the use of a FlowCam instrument, access to VisualSpreadsheet software, and a full suite of training modules, many students present papers and posters at national or international conferences. One such example is Zabdiel Roldan Ayala, who was a graduate student studying phytoplankton at Queens College's School of Earth and Environmental Sciences. Zabdiel used a FlowCam 8100 instrument during the Summer of 2021 to observe and record phytoplankton community trends in Long Island Sound.
Supported by Yokogawa Fluid Imaging, Zabdiel presented his research at the Virtual 2022 Ocean Sciences Meeting, an annual conference co-sponsored by the American Geophysical Union (AGU), the Association for the Sciences of Limnology and Oceanography (ASLO), and The Oceanography Society (TOS). His talk was titled "Applications of the FlowCam 8100 for Characterizing Phytoplankton Communities in an Urban Estuary During Summer of 2021"
Here is an expert from Zabdiel's abstract:
"Long Island Sound (LIS) receives high levels of anthropogenic nitrogen (N) inputs from the nearby New York City (NYC) area due to combined sewer overflow systems. Despite known associations between nitrogen and harmful algal bloom development, the extent to which LIS phytoplankton community composition varies across spatial and temporal scales remains less characterized. The FlowCam 8100 [can help] resolve these knowledge gaps because of its unique ability to develop phytoplankton image libraries to identify species, calculate cell biovolume, and count particles."

Zabdiel used FlowCam to classify organisms from his Western Long Island Sound samples, including Prorocentrum, pictured above. Since presenting his grant research findings at the Ocean Sciences Meeting, his passion for studying HABs has continued to evolve. Keep reading to learn about his most recent publication: A comparison between the FlowCam8100, microscopy, and sandwich hybridization assay for quantifying abundances of the saxitoxin-producing dinoflagellate, Alexandrium catenella.
Several methods have been developed to monitor harmful algal bloom (HAB) species. Light microscopy is commonly used due to its widespread availability and ability to connect new findings with historical data. However, microscopy can be time-consuming when analyzing individual samples and requires taxonomic expertise, making it challenging to differentiate between morphologically similar species.
 Flow Imaging Microscopy, specifically via FlowCam, integrates flow cytometry and microscopy principles to capture, identify, and electronically quantify particles in a fluid medium. The FlowCam 8100 instrument has been utilized in various research studies, including those focusing on microplastics, lipids, cell biovolume, and phytoplankton. FlowCam offers a more efficient analysis process compared to traditional microscopy, albeit potentially with slightly reduced taxonomic resolution and cell concentrations. (Pictured here, Alexandrium catenella as imaged by FlowCam)
Flow Imaging Microscopy, specifically via FlowCam, integrates flow cytometry and microscopy principles to capture, identify, and electronically quantify particles in a fluid medium. The FlowCam 8100 instrument has been utilized in various research studies, including those focusing on microplastics, lipids, cell biovolume, and phytoplankton. FlowCam offers a more efficient analysis process compared to traditional microscopy, albeit potentially with slightly reduced taxonomic resolution and cell concentrations. (Pictured here, Alexandrium catenella as imaged by FlowCam)
Sandwich hybridization assay (SHA) is another technique that enables rapid (<1 h) taxon or species-specific plankton identification and quantification using ribosomal RNA (rRNA)-targeted oligonucleotides and has been applied to several HAB species. Comparisons between qPCR, SHA, and microscopy, have demonstrated strong agreement for HAB cell quantification.
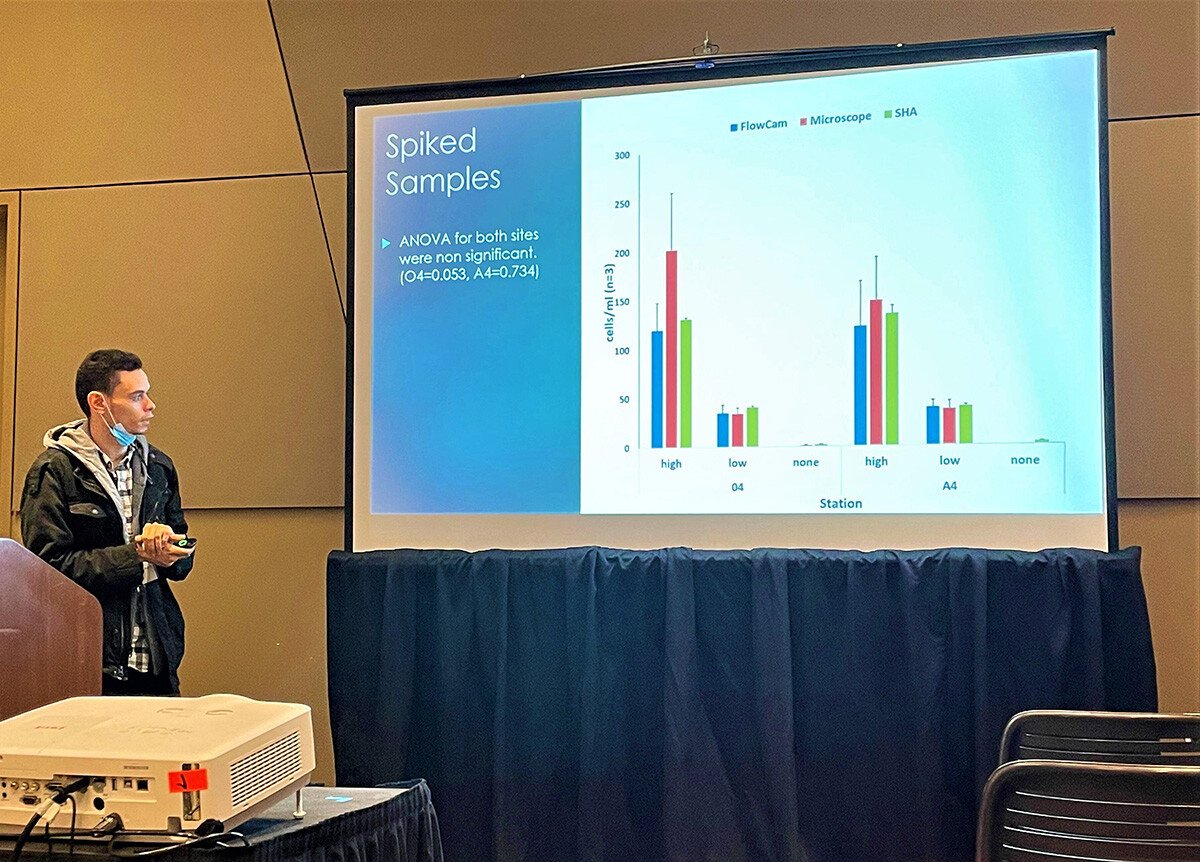
Each approach (microscopy, FlowCam, SHA) has been used for HAB detection and monitoring. Still, they have not been uniformly compared despite marked differences regarding equipment, quantification mechanism, cost, training, and other attributes. Given the increasing emphasis on networking technologies to enhance bloom warnings, reconciling datasets generated by these tools is a critical management need. This study focused on the saxitoxin-producing 'red tide' dinoflagellate Alexandrium catenella because it causes paralytic shellfish poisoning, has an expansive biogeographic distribution, and is monitored worldwide. The authors present data comparing the three different techniques and suggest that the results would be broadly applicable to several HAB species.
Read the full paper: